Protein-Protein Docking and Molecular Dynamics Simulations Elucidated Binding Modes of FUBI-p62 UBA Complex
Abstract
The cytosolic Fau protein, a precursor of antimicrobial peptide, composes of
ubiquitin-like domain FUBI at N-terminus and ribosomal protein rpS30 at C-terminus. Fau
has been important in killing of intracellular Mycobacterium tuberculosis infection through
autophagy-targeting p62 mechanism. The p62 adapter protein delivered microbicidal protein
rpS30 to autolysosome where it was converted into antimicrobial peptides capable of killing
M. tuberculosis in mycobacterial phagosome. Recently, direct interaction of FUBI and p62
UBA domain has been established using immunoprecipitation. In the absence of experimental
complex structure of FUBI and p62 UBA, understanding of binding interaction could be
extensively characterized using molecular modeling techniques. The aim of this study was to
elucidate the binding mode of FUBI interacting with the p62 UBA. Base on the conserved
hydrophobic binding regions of FUBI and p62 UBA domain, 334 docked poses were
predicted using the ZDOCK and RDOCK protein-protein docking algorithms. Five binding
modes of complex structures were clustered and only two were stable after 15 ns of
molecular dynamics simulations. The binding free energy was elucidated using MM-PBSA
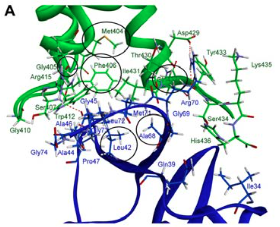
method and the best FUBI-p62 UBA complex was determined. The key binding residues of
FUBI and p62 UBA domain were elucidated using protein interface and alanine scanning.
Gly405 and Phe406 in the MGF hydrophobic area and certain residues in loop 1, helix 2 and
helix 3 of the p62 UBA domain were bound with FUBI domain. The results enable us to
understand how p62 and Fau interact which is a crucial step of autophagy-targeting p62
mechanism for antimycobacterial action.
Full Text:
171-179:PDFRefbacks
- There are currently no refbacks.

